Real Time Analytics to Monitor and Predict Emerging Plant Disease
 We are excited to announce our new Predictive Intelligence for Pandemic Preparedness (PIPP) Grant titled “Real-Time Analytics to Monitor and Predict Emerging Plant Disease” was awarded for $1 Million USD by the National Science Foundation.
We are excited to announce our new Predictive Intelligence for Pandemic Preparedness (PIPP) Grant titled “Real-Time Analytics to Monitor and Predict Emerging Plant Disease” was awarded for $1 Million USD by the National Science Foundation.
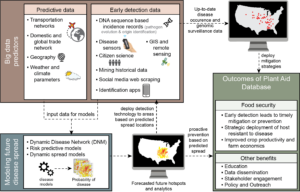
Plant disease outbreaks are increasing and threaten food security for the vulnerable in many areas of the world and in the US. A stable, nutritious food supply is needed to both lift people out of poverty and improve health outcomes. Plant diseases cause crop losses from 20 to 30% in staple food crops. Plant diseases, both endemic and recently emerging, are spreading and exacerbated by climate change, transmission with global food trade networks, and emergence of new strains that may be difficult to control. This team of researchers will develop better ways to detect and predict when and where plant diseases will emerge. This research will characterize how human attitudes and social behavior of stakeholders impacts plant disease transmission and adoption of sensor, surveillance and disease prediction technologies.
Prediction of plant disease pandemics is unreliable due to the lack of real-time detection, surveillance, and data analytics to inform decision-making and prevent spread. This is the grand challenge that the convergence research team will tackle in this Predictive Intelligence for Pandemic Prevention (PIPP) planning grant. In order to improve pandemic prediction and tackle this grand challenge, a new set of predictive tools are needed. In the PIPP Phase I project, the multidisciplinary team will develop a pandemic prediction system called the “Plant Aid Database (PAdb)” that links pathogen detection by in-situ plant disease sensors and remote sensing of crop health, genomic surveillance, real-time spatial and temporal data analytics and climate data to predictive simulations of plant disease pandemics. The team plan to validate the PAdb using several model plant pathogens including novel lineages of Phytophthora infestans and the cucurbit downy mildew pathogen Pseudoperonospora cubensis.
The team engaged a broad group of stakeholders including scientists, growers, extension specialists, the USDA APHIS Plant Protection and Quarantine personnel, the Department of Homeland Security inspectors, and diagnosticians in the National Plant Diagnostic Network in a Pandemic Preparedness Symposium, poster session and workshop held at NC State on April 5-6, 2023. Stakeholder experiences and perspectives on detection, response and mitigation of pathogens was examined using current methods and the PAdb was introduced for discussion. The team also engages a diverse group of postdoctoral associates, graduate students and research staff through research and workshop participation and foster partnerships for a future Plant Disease Pandemic Preparedness Center.
Ristaino, J. B. , Delborne, J., Zering, K., Jones, C., Tateosian, L., Vatsavai, R., Ojiambo. P., Carbone, I, Meentemeyer, R., and Wei, Q. 2021. Real-Time Analytics to Monitor and Predict Emerging Plant Diseases. NSF PIPP, $1,000,000, 8/1/22-12/31/23.
Select Recent NSF PIPP Team Publications
- Coomber, A., Saville, A.C., Martin, M., Carbone, I., and Ristaino, J. 2024 A pangenome analysis reveals center of origin and evolutionary history of the Phytophthora Ic clade. Plos One, accepted with revisions.
- Mainello-Land, A., Saville, A.C., Acharya, J., and Ristaino, J. 2024. Loop-mediated isothermal amplification to detect Phytophthora kernoviae, P. ramorum, and the P. ramorum NA1 using lyobeads and a smartphone platform. Phytopathology: in press.
- Coomber, A., Saville, A.C., and Ristaino, J. 2024. Evolution of Phytophthora infestans on its Solanum host since the Irish potato famine. Nature Communications 15:6466.
- Shymanovich, T., Saville, A.C. Paul, R., and Ristaino, J. 2024. Rapid detection of viral, bacterial, fungal, and oomycete pathogens on tomato with microneedles, LAMP on a microfluidic chip, and smartphone device. Phytopathology First Look.
- Tateosian, L., Saeffer, A., Yang, Y. P., Saville, A., and Ristaino, J. B. 2024. Reconstructing 19th century and modern potato late blight outbreaks using text analytics. Nature Scientific Reports 14: 2523.
- Shymanovich, T, Saville, A, Mohammed, N, Wei, Q., Rasmussen, D, Lahre, K., Rotenberg, D., Whitfield, A., and Ristaino JB. 2024. Disease progress and detection of the tomato spotted wilt virus resistance breaking strain with LAMP and CRISPR assays. PhytoFrontiers 4: 50-60.
- Coomber, A., Saville, A.C., Carbone, I. and Ristaino, J. 2023. An open access T-BAS phylogeny for emerging Phytophthora species. Plos One: 18(4): e0283540
- Sanchez, F., Galvis, J.A., Cardenas, N.C., Corzo, C., Jones, C., and Machado, G. 2023. Spatiotemporal relative risk distribution of porcine reproductive and respiratory syndrome virus in the United States. Frontiers in Veterinary Science 10:1158306.
- Saville, A., McGrath, M, Jones, JC., Polo, J. and Ristaino, J. 2023. Understanding the genotypic and phenotypic structure and impact of climate on Phytophthora nicotianae outbreaks on potato and tomato in the eastern US. Phytopathology 113:1506-1514.
- Lee, G., Hossain, O. , Jamalzadegan, S., Liu, X., Wang, H., Saville, A. C., Shymanovich, T., Paul, R., Rotenberg, D., Whitfield, A. E., Ristaino, J. B., Zhu, Y., and Wei, Q. 2023. Abaxial leaf surface-mounted multimodal wearable sensor for continuous plant physiology monitoring. Science Advances: 9 eade2232.
- Tateosian, G. Saffer, A, Walden-Schreiner, C., Shukunobe, M. 2023. Plant pest invasions, as seen through news and social media. Comput. Environ. Urban Syst. 100: 101922.
- Weng, Z., You, Z., Yang, J., Mohammad, N., Lin, M., Wei, Q., Gao, X., Zhang, Y. 2023. CRISPR-Cas biochemistry and CRISPR-based molecular diagnostics. Angewandte Chemi DOI: 10.1002/anie.202214987
- Gugino, B. K., Britton, W., Keinath, A. P., McGrath, M. T., Melanson, R. A., Miller, S. A., LaForest, J. H. and Ojiambo, P. S. 2022. Cucurbit downy mildew ipmPIPE: A valued resource for information dissemination and in-season disease management decisions for diverse stakeholders. Phytopathology 112:163.
- Jones, C., Skrip, M., Seliger, B., Jones. S., Wakie, T., Takeuchi, Y., Petras, V., Petrasovea, A., and Meentemeyer, R. 2022. Spotted lanternfly predicted to establish in California by 2033 without preventative management. Comm. Biol. 5:558.
- Kikway, I., Keinath, A.P., and Ojiambo, P.S. 2022. Temporal dynamics and severity of cucurbit downy mildew epidemics as affected by chemical control and cucurbit host type. Plant Disease 106: 1009-1019.
- Paul, R., Ostermann, E., Wei, Q. 2022. Rapid extraction of plant nucleic acids by microneedle patch for in-field detection of plant pathogens. In: Luchi, N. (eds) Plant Pathology. Methods in Molecular Biology, vol 2536. Humana, New York, NY.
