An Open T-BAS Phylogeny for Emerging Phytophthora’s
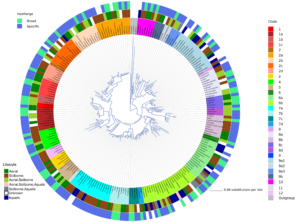
Phytophthora species cause severe diseases on food, forest, and ornamental crops. Since the genus was described in 1875, it has expanded to comprise over 190 formally described species. There is a need for an open access bioinformatic tool that centralizes diverse streams of sequence data and metadata to facilitate research and identification of Phytophthora species. We used the Tree-Based Alignment Selector Toolkit (T-BAS) to develop a phylogeny of 192 formally described species and 33 informal taxa in the genus Phytophthora using sequences of eight nuclear genes. The phylogenetic tree was inferred using the RAxML maximum likelihood method. The T-BAS tool provides a visualization framework allowing users to place unknown isolates on a curated phylogeny of all Phytophthora species. Critically, this resource can be updated in real-time to keep pace with new species descriptions. The tool contains metadata such as clade, host species, substrate, sexual characteristics, distribution, and reference literature, which can be visualized on the tree and downloaded for other uses. This phylogenetic resource will allow data sharing among research groups and the database will enable the global Phytophthora community to upload sequences and determine the phylogenetic placement of an isolate within the larger phylogeny and to download sequence data and metadata. The database will be curated by a community of Phytophthora researchers and housed on the T-BAS web portal in the Center for Integrated Fungal Research at NC State. The T-BAS web tool can be leveraged to create similar metadata enhanced phylogenies for diverse populations of pathogens.
Access tree placement tutorial
Access the Phytophthora T-BAS tool
Coomber, A., Saville, A., Carbone, I. and Ristaino, J. 2023. An open T base phylogeny for emerging Phytophthora species. Plos One: https://doi.org/10.1371/journal.pone.0283540
For questions on the use of the tool or on submission of new species, please contact Jean Ristaino, Allison Coomber, or Amanda Saville.
For technical issues, please contact Ignazio Carbone
